Where global policy meets interoperability: the thorny issue of digital sequence information
January 4, 2023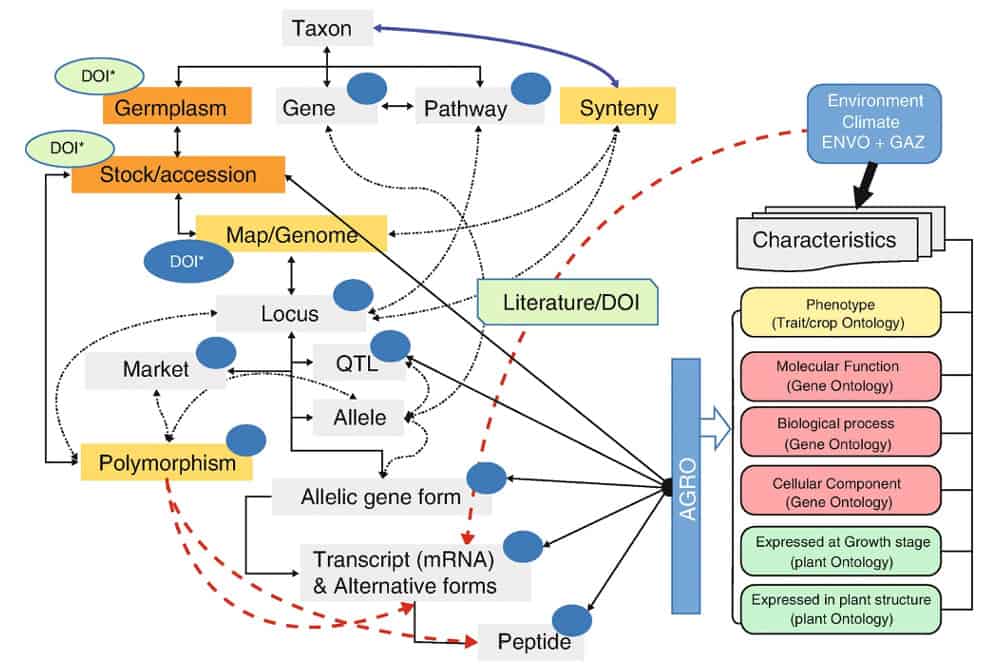
Figure 1. Schematic showing the utility of digital genetic objects (DGOs), represented with dark blue circles. These knowledge objects can be assigned distinct types of digital sequence information (DSI), and associated with digital object identifiers (DOIs), allowing for greater interoperability between data systems. (Source: Manzella et al. 2023)
Corresponding author: Pankaj Jaiswal, Oregon State University, Corvallis, OR, USA.
In October last year, a team of experts from FAO, CGIAR and Oregon State University published an Open Access book chapter titled ‘Digital Sequence Information and Plant Genetic Resources: Global Policy Meet Interoperability.’
The chapter appears in ‘Towards Responsible Plant Data Linkage: Data Challenges for Agricultural Research and Development’, alongside many other insightful perspectives by leading experts in data curation and governance.
The chapter takes stock of the information management systems and global policy frameworks that currently regulate plant genetic resources (PGR). In particular, they examine the status of digital sequence information (DSI) – a contentious issue for policy makers and genomic science communities alike.
DSI is at the heart of modern bioscience. “With the deluge of reference and pan-genome sequence available, researchers have very quickly identified large volumes of SNPs and other genetic markers,” explains Pankaj Jaiswal, one of the authors of the chapter.
“These markers are now available for studying genetic diversity, genotype-to-phenotype association studies, integrating sequences with improved traits in plant breeding materials, as well as selecting candidate genes for genetically engineering functional gains, like improving yield or boosting stress resilience.”
The strategies that utilise plant genetic DSI (such as accelerated breeding and CRISPR-based approaches) are helping to address numerous global food security challenges.
On the one hand, F.A.I.R (Findable, Accessible, Interoperable, Reusable) access to nucleotide and protein sequence information (DSI) is guiding and accelerating these developments. But on the other hand, DSI has the potential to undermine some of the global policy frameworks designed to ensure that benefits from agricultural science are equitably shared.
The international regulations on access and benefit-sharing (ABS) from the use of plant genetic resources include the International Treaty on Plant Genetic Resources for Food and Agriculture (The Treaty) and the Nagoya Protocol of the Convention on Biological Diversity (CBD).
At present, these ABS systems focus on regulating access to the physical, tangible components of genetic resources, i.e. germplasm. “Increasingly, global discussions are taking place on whether to regulate the intangible components (i.e. DSI) within the remit of these agreements,” says Jaiswal.
In their chapter, the authors provide an insightful overview of current discussion on DSI and ABS policies up to the fifteenth Conference of the Parties to the CBD (COP 15), which was held shortly after the chapter was published. They then weave a compelling argument that in order to implement any future legal solutions, we must first enhance the interoperability of relevant data systems.
The authors go on to propose actionable solutions for associating DSI with its source germplasm, enabling reciprocal citations and data exchange between repositories of source material and repositories of genomic data.
Firstly, they propose integrating the federated system of databases of the International Nucleotide Sequence Database Collaboration (INSDC) through the Treaty’s Global Information System (GLIS) via the well-known mechanism of digital object identifiers (DOIs).
“DOIs are an ideal foundation because they have flexible metadata structures,” explains Jaiswal. “[This] allows for descriptions specific to each type of knowledge object – from publications, to gene bank accessions, to datasets, to genetic markers and so on.”
“GLIS already gives researchers the option to assign DOIs to their plant genetic resources” says Jaiswal. The authors suggest that genomics repositories like INSDC could adopt a standard of citing DOIs attributed to specific source material – whether assigned by GLIS, or by another authority. Of course, this will require discussing global metadata standards for DOI-based DSI.
Secondly, the authors propose the use of digital genetic objects (DGOs) to address some of challenges arising from the messy ambiguity of what constitutes DSI. Since there are so many different types of information (e.g. nucleic acid sequence reads, data about gene expression and function, unsequenced markers and chromosomal segments), they suggest assigning a DGO to every distinct piece of DSI. Each of these DGOs can be linked to the DOI of the source material, and thus facilitate discovery via GLIS and interoperability among genomic data repositories.
A major roadblock in implementing these solutions would be user motivation to adhere to new standards – which is why pilot projects, community discussion, training sessions, and raising awareness of the benefits of interoperability is a must. “The concept has far-reaching impacts, not just for the plant community, but across life sciences, and would enable better citation indexing, provenance and benefit sharing,” says Jaiswal.
We encourage all of our members to read this thought-provoking book chapter to deepen their understanding of the opportunities for greater data linkage in the world of plant genetic resources, and the benefits this could bring to all stakeholders.
References:
Manzella, D., Marsella, M., Jaiswal, P., Arnaud, E., King, B. (2023). Digital Sequence Information and Plant Genetic Resources: Global Policy Meets Interoperability. In: Williamson, H.F., Leonelli, S. (eds) Towards Responsible Plant Data Linkage: Data Challenges for Agricultural Research and Development. Springer, Cham. https://doi.org/10.1007/978-3-031-13276-6_10
Back
Recent Comments