Meet Tripal, an open toolkit for building biological databases
October 12, 2022Written by Lacey Sanderson, University of Saskatchewan for publication in DivSeek Connect, Issue 3.
Tripal is a toolkit for developing web data portals focused on supporting open-science by helping researchers build sites for community engagement, distributing scientific data and providing domain-specific tools for analysis.
Tripal serves the needs of a diverse community with one core developer focused on primary genomic research and the other focused on breeding and applied agriculture research.
This has resulted in a very generic core with (1) an ontology-focused approach to data storage, (2) a unified interface for data curation, analysis and exploration focused on data attribution and (3) a built-in system for providing layers of access for data security while facilitating collaboration.
Underlying all core functionality is an extensive and robust set of APIs allowing site developers to customize and extend every aspect of their data portal.
An example of this is KnowPulse, which is a Tripal data portal focused on germplasm diversity data in support of the Pulse Crop Breeding program at the University of Saskatchewan.
Through open-source modules available to all Tripal sites, KnowPulse provides germplasm-focused tools for exploring large-scale phenotypic and genotypic data in multiple ways.
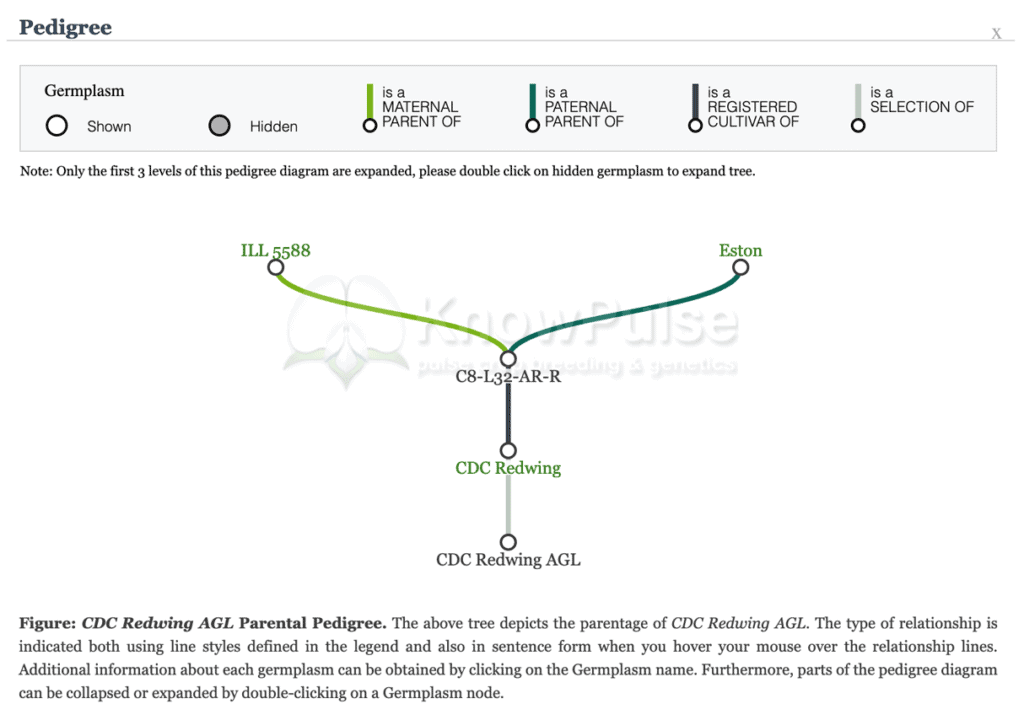
Screenshot from the Tripal data portal KnowPulse, showing pedigree overview for an accession of lentil (Lens culinaris). (Source: Lacey Anderson, University of Saskatchewan).
Wherever possible, tools are embedded directly in the site to reduce activation energy for breeders looking into new technologies and highly couple the data and tools for a seamless, interconnected experience.
For example, on a germplasm accession page, there is important metadata and attribution right at the top, followed by a graphical pedigree to provide context and fully describe the germplasm concisely.
All data available for that germplasm is summarized and tools linked or embedded directly. For example, an embedded violin plot highlights the phenotype of the current accession within the greater context of various experiments. This allows breeders to answer important questions at a glance (e.g. is this accession early flowering in our diversity panel for adaptation in our growing zone?).

Screenshot from the Tripal data portal KnowPulse, showing phenotype overview for an accession of lentil (Lens culinaris). (Source: Lacey Anderson, University of Saskatchewan).
The open-source nature of Tripal supports every community to focus on their individual needs while making it easy for them to produce extensions for others at the same time.
This approach has helped Tripal thrive for 10 years with over 125 current sites world-wide and an active community including advisory and project management committees open to all members.
Corresponding authors: Lacey Anderson ([email protected]) and Kirstin Bett ([email protected]) of the University of Saskatchewan
Back
Recent Comments